I use mathematical modelling techniques to model infectious disease spread within the body, employing a variety of advanced methods to fully capture the complexity of biological systems. Specifically, I utilise differential equations and individual-based models to mimic disease progression and simulate novel treatment strategies, taking into account various factors such as population dynamics, immune responses, and environmental influences. Through these models, I can analyse the impact of different interventions, predict potential outcomes, and ultimately contribute to the development of more effective treatment protocols that can improve patient outcomes and public health strategies.
Tuberculosis (TB)
I am currently focused on modelling tuberculosis (TB) infection, using modelling to help to improve treatment strategies and understand more about disease relapse, which is a critical aspect of managing this pervasive health challenge. This research is particularly important as TB remains a leading cause of mortality worldwide, and its complex nature requires innovative approaches to treatment and prevention.
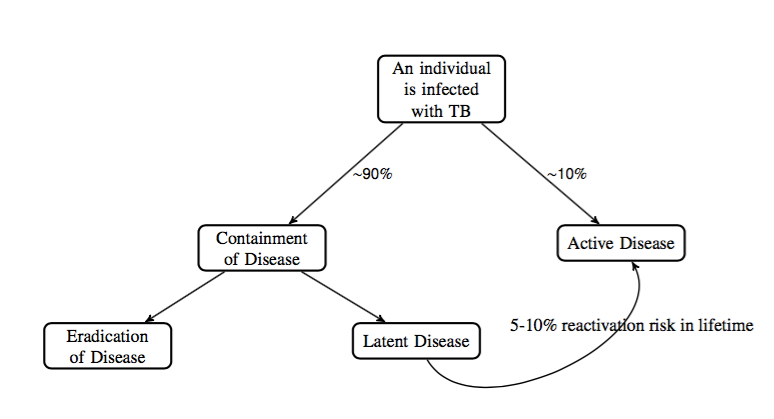
Schematic of population percentages of outcomes of TB disease
With support from my Medical Research Council (MRC) fellowship, I will enhance my mathematical model to enable virtual testing of multiple antibiotics simultaneously, thereby allowing for a more comprehensive exploration of treatment strategies. This integration will be executed via an advanced pharmacokinetic/pharmacodynamic (PK/PD) model, which will facilitate a deeper understanding of drug interactions and their effects on the TB pathogen. We will also extend the model to account for common co-morbidities such as diabetes and HIV. Ultimately, this work aims to contribute valuable insights that could inform future clinical trial design and improve patient outcomes in the fight against tuberculosis.

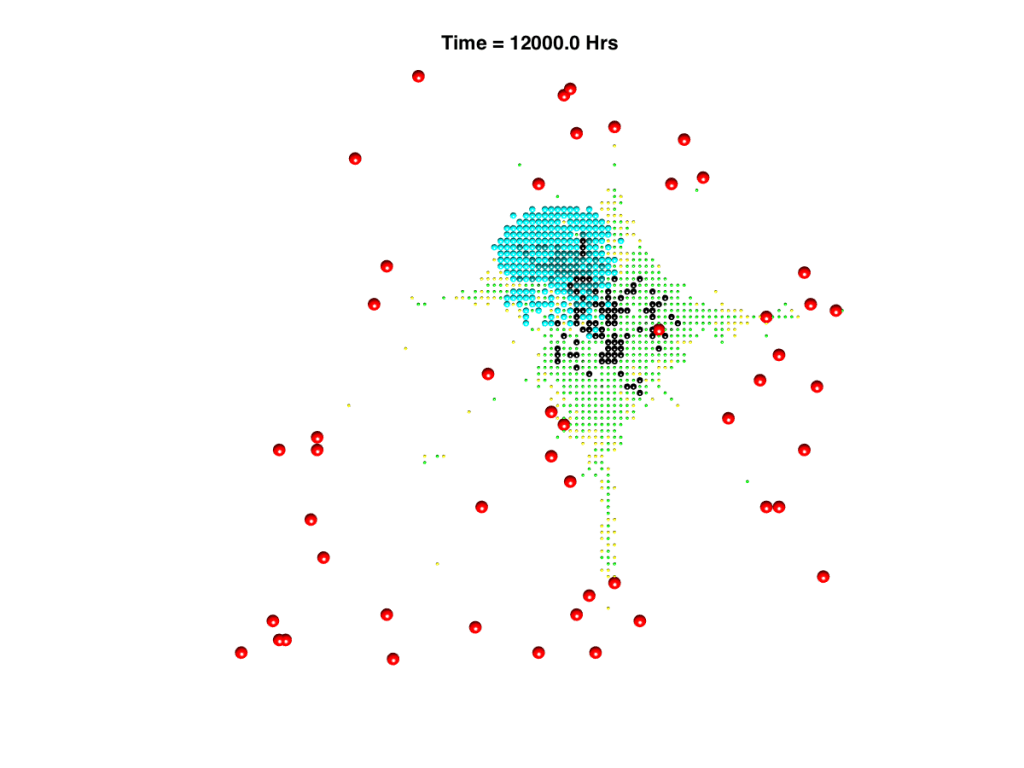
Example spatial plots from individual-based model. Left hand plot shows a concentration profile of oxygen supplied by the blood vessel network, right hand plot shows an example simulation outcome of a TB infection in a small area of the lung
COVID-19
The group and I have been using similar modelling techniques to try to understand more about this serious disease. Thanks to a Chief Scientific Office grant that we were awarded in 2020, Chris Rowlatt and I have been working with our collaborators to develop a within-host mathematical model of SARS-CoV-2 infection. In 2021 I was awarded a RAMP Early Career Investigator Award for this work. See our pre-print https://doi.org/10.1101/2022.05.06.490883 on our model. We’re also really grateful to have had funding from EPSRC to continue with this work.
Antimicrobial Resistance (AMR)
Resistance to antibiotics is one of the most imminent threats to public health worldwide. The World Health Organization has recognized antimicrobial resistance (AMR) as “a global health security threat that requires action across government sectors and society as a whole.” New resistance mechanisms are emerging and spreading, threatening our ability to treat common infectious diseases, resulting in prolonged illness, disability, and death. Without effective antimicrobials for prevention and treatment of infections, medical procedures such as organ transplantation, cancer chemotherapy, diabetes management and major surgery become very high risk. Infections that are currently simple to treat could be fatal. Furthermore, AMR increases the cost of health care and results in lengthier hospital stays.
I was fortunate enough to be awarded a Springboard Award in 2018 from the Academy of Medical Sciences to complete some work on modelling resistant forms of urinary tract infections along with a previous PhD student. See the group publication list for the outcomes of this work.